Cellular and extracellular non-coding RNA in heterogeneous brain tumor microenvironment.
While significant effort has been dedicated to identify sequence and expression alterations of protein-coding genes, contributing vastly to our understanding of brain tumor; little is known about functional consequences of alterations in non-protein coding genes (ncRNAs). Additional layer of complexity are microenvironmental rearrangements contributing to intra-tumoral heterogeneity inducing abnormalities/alteration of gene expression. Such phenotypical diversity can be rendered even more complex by the inter-cellular transfer of functional molecules encapsulated in extracellular vesicles (EVs, including exosomes).
The complexity of solid tumors, including GBM, and their distinct pathophysiology, relies on anatomic niches that transmit and receive signals through cellular and acellular mediators. A lack of experimental models of tumor heterogeneity limits our knowledge of the complex subpopulation dynamics within the tumor ecosystem which undergo constant architectural, phenotypic, and transcriptomic re-arrangement depending on fluctuating microenvironmental contexts as the disease progresses. Using clinically relevant, patient-derived cancer cells and novel heterogeneous xenograft tumor model we elucidate whether the biological mechanisms by which selected tumor cells acquire the ability to adapt and survive under severe conditions are of significant clinical importance.
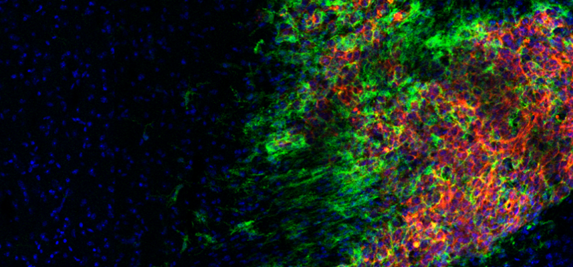
EVs carry multiple molecules, delivery of whose may reprogram recipient cells either through passive delivery of molecular cargo, and/or by active regulation of transcriptional profiles. Both mechanisms can contribute to the modification of the tumor microenvironment and enhanced tissue and cellular heterogeneity. Our goal is to uncover GSC-EV cargo in the context of distinct transcriptome subclasses to provide evidence for functional role of EVs and their cargo in maintaining such diverse landscapes.
To date, analyses of molecular diversity have focused mostly on protein-coding genes and the role and contribution of ncRNAs (including long, non-coding RNA – lncRNA) which constitute a vast majority of the human transcriptome, remains insufficiently studied and characterized in GBM. The primary goal of this study is to evaluate the capability of ncRNAs as novel GBM drivers. Subsequent goal is to use ncRNAs as putative therapeutic targets. Thus in this project we aim to understand the mechanisms of ncRNA regulation in heterogeneous populations of GBM stem cells (GSCs) in the context of brain tumor microenvironment and the role of ncRNAs in controlling subtype-specific adaptation of GSC to environmental challenges including hypoxia, to define the ncRNA protein interactome and to explain how ncRNAs achieve their regulatory specificity by their downstream effectors.